About our work
The goal of the Savage Lab is to understand how protein machinery facilitates the biochemistry of the cell. We are particularly interested in photosynthetic CO2 fixation, a complex set of reactions that must occur in the right place and time to capture and convert light into stored chemical energy. Photosynthesis is uniquely important for human society. It catalyzes the assimilation of inorganic carbon into the organic world and is the ultimate source of nearly all nutritional calories on Earth.
We employ several model organisms including various bacterial species, mammalian cells and plant cells. For the past several years, we have focused much of our effort on the bacterial Carbon dioxide Concentrating Mechanism (CCM), a multi-scale, multi-component system that functions to increase the rate and specificity of the critical enzyme ribulose-1,5-bisphosphate carboxylase / oxygenase (Rubisco). We use the tools of biochemistry, molecular biology, and synthetic biology to identify and characterize the key players and mechanistic principles underlying CCM function. Ultimately, we hope to develop a total cell biological understanding of how CCM function emerges from individual components and to use this understanding for the improvement of photosynthetic CO2 fixation in plants.
We are therefore also interested in the development of novel genome editing technologies for accelerating genetic experiments in plants. Given the generality of these tools, we also explore how engineered genome editors can be used to improve editing in human cells. Our specific areas of research are as follows.
Towards a Total Cell Biological Understanding of CO2 Concentrating mechanisms
Systems model of the CCM adapted from Mangan et al. 2016
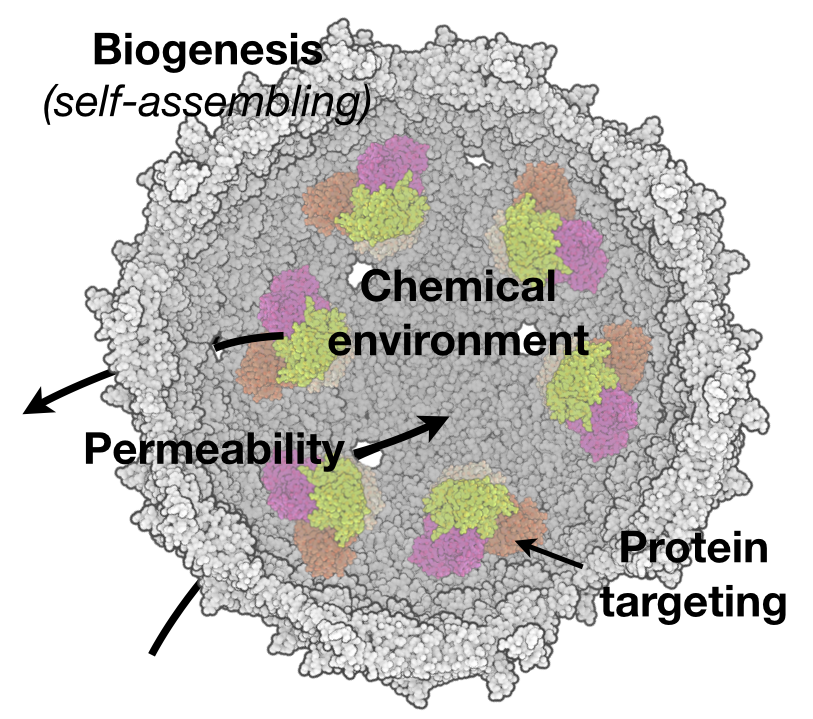
Cyanobacteria carry out photosynthetic carbon dioxide assimilation by controlling when and where these biochemical reactions occur. Perhaps most amazing is the use of a protein organelle, the carboxysome, to achieve improved activity of Rubisco. Rubisco is a notoriously bad enzyme, and it is thought the inside of the carboxysome provides a microenvironment within the cell of high carbon dioxide (and possibly low oxygen) as a means of improving rate and specificity. Despite numerous structural studies of carboxysome components, there are many open questions we are currently investigating related to how this massive complex self-assembles and functions in the context of the cellular milieu. For example, our systems model of the CCM (Mangan et al. 2016) suggests that pH, carbon transport, and carboxysome permeability are highly intertwined and we are interested in mechanisms that can control the permeability of the carboxysome shell. Second, work from our group has demonstrated the inherent genetic modularity of the CCM may all for it to be transferred to other organisms in order to improve their growth (Bonacci et al. 2012). We are therefore also developing tractable genetic systems to explore, in a synthetic biological context, how CCMs can be constructed de novo. We have used high-throughput screening strategies to identify unknown components of the CCM (Desmarais et al. 2019), and in a recent paper, demonstrated a complete reconstitution of the CCM by porting it from a native to a heterologous host (Flamholz et al. 2020). The latter publication is the first such successful demonstration of a fully functional, engineered CCM.
Genome Editing Technology development
Insertional profiling of functional hotspots in the Cas9 protein (red denotes enriched, blue depleted) from Oakes et al. 2016.
An important application of our protein engineering work is to make robust genome editing that can be delivered to the right cell at the right time. For example, the study of many model organisms, particularly plants, is constrained by our ability to quickly and easily manipulate gene sequence and gene expression. Although advances with CRISPR-Cas proteins, such as Cas9, have accelerated this progress, much remains to be done. To this end, we have developed engineered variants of Cas9 that are allosterically regulated (Oakes et al. 2016). More broadly, we have sought to optimized the entire scaffold of Cas9 using topological mutagenesis in order to improve both delivery (Oakes et al. 2019) and for effector fusions such as base editors (Huang et al. 2019). In more recent work, we have also developed a technique, called MISER, that enables the directed evolution of smaller genome editing proteins which are more suitable for viral delivery vectors (Shams et al. 2021).
Tools for Protein Engineering
Domain insertion libraries can be used to engineer proteins with highly novel functions, with a variety of downstream applications.
Our group takes an integrative view of protein function. That is, we are interested not only in protein mechanism but also in trying to understand this mechanism in its native, cellular context. We therefore also develop genetic tools to interrogate proteins in the cell. Much of this effort has been focused on leveraging recent advances in DNA sequencing to interrogate protein function in higher throughput and also demonstrating that protein sequence topology is a critical, yet often unexplored, dimension of the sequence-function landscape (Higgins and Savage 2017). Highlights of our work include developing a novel systematic mutagenesis method (Higgins et al. 2017), a transposon-based method for rapidly constructing metabolite biosensors (Nadler et al. 2016), and development of an allosterically regulated Cas9 variant (Oakes et al. 2016). In more recent work, we have also developed a technique, called MISER, that enables the directed evolution of smaller proteins from a starting parental scaffold (Shams et al. 2021). Currently, we are developing high-throughput selective assays for some of our favorite enzymes, including Rubisco (Flamholz et al. 2020), and applying modern mutational methods and machine learning approaches to better explore sequence-function landscapes.
Understanding the Diversity and Functions of Protein Compartments
The carboxysome is a 250 MDa+ complex formed from thousands of proteins, yet it self-assembles in the bacterial cytosol into monodisperse particles. How does this amazing process occur? In previous work, we have used microscopy to demonstrate the importance of carboxysome partitioning during cell division and the surprising connection between this process and the bacterial cytoskeleton (Savage et al. 2010; Yokoo et al. 2015). More recently we have focused on assembly directly and have demonstrated the minimal set of proteins sufficient to produce the carboxysome in a heterologous host (Bonacci et al. 2012), and honed in on one of these proteins, CsoS2, as particularly critical to the process (Chaijarasphong et al. 2016). We are now currently building on these advances to understand how specific protein-protein interactions direct the assembly process and are attempting to elucidate the fundamental biophysical principles leading to high fidelity assembly. Finally, we now believe that the carboxysome is yet one example of myriad ways in which protein compartments can facilitate biochemical function (Nichols et al. 2017). We are thus also interested identifying novel compartments and, more broadly, understanding the functional principles of compartmentalization. An example of the former is our recent identification and characterization of a wide variety of compartments involved in sulfur metabolism (Nichols et al. 2021).
Note that plasmid reagents from the Savage Lab are available from our addgene site and that all source code from our work is available on our github site.
About Cal
The Savage Lab is a member of the Department of Molecular & Cell Biology at the University of California, Berkeley. UC Berkeley, also affectionately known as Cal, is a public research university dedicated to both education and research and has made such varied contributions to society as the discovery of six chemical elements and the birth of the Free Speech Movement. Cal is located near downtown Berkeley, California and is rooted in the vibrant intellectual, cultural, environmental, culinary, and entrepreneurial communities of the San Francisco Bay area.
The Savage Lab is also a member of the Innovative Genomics Institute (aka the IGI), a collaboration between UC Berkeley and UC San Francisco. This collaboration of diverse expertise seeks to develop genome editing technology and apply it for the public good in biomedical science and agriculture. The IGI is located in the IGI Building (IGIB), a 113,000 square foot research building on the northwest corner of campus adjacent to downtown Berkeley. The IGIB houses laboratories from numerous departments on campus and possesses state-of the art research facilities.